Reading the Skywater130 raw data
Google released raw experimental data for the SKY130 open source process technology. This data is stored in the MDM format which is ideal to show the application of DMTs data acquisition tools. A simple direct read of a single measurement to a DataFrame is shown in the test case test_read_skywater_mdm
In this example, we will show how to read all devices in the measurement data into a DMT.DutLib. Also, it is shown how to access the data inside the library and finally how to document the measurement in LaTeX.
The Skywater read in code
""" Reading the skywater130 data into a DMT DutLib
This large example shows how to read in all the skywater130 pdk measurement data into a DMT DutLib object. The data processing can also be done in Python. Feel free to use this example as a starting point for bulk reading of data.
Before you can use this data, you have to clone the measurement data repository from:
https://github.com/google/skywater-pdk-sky130-raw-data
This example is used for the initial Skywater 130 data from July 2022.
The file structure of the Skywater PDK does not fit directly to the DMT structure. DMT assumes that 1 folder holds data of one physical device.
Since this is not the case for the skywater data, we will have to rearrange the measurement data.
So, in the first step the raw data is sorted so that every folder holds data for one physical device.
"""
import shutil
import re
import numpy as np
from pathlib import Path
from DMT.core import DutMeas, DutType, DocuDutLib, DutLib, specifiers, sub_specifiers, Plot
path_to_dmt_core = Path(__file__).resolve().parent.parent.parent.parent
# assume Skywater data is at this location relative to DMT-core:
path_to_git = path_to_dmt_core.parent.parent / "skywater-pdk-sky130-raw-data-main"
path_to_cells = path_to_git / "sky130_fd_pr" / "cells"
# sort the measurement data files into folders using shutil
for child in path_to_cells.glob("*/" * (2)):
if (
child.is_file()
): # only files are allowed since directories are created this does not interfere
device_name = child.stem[: child.stem.find("_ID")]
target_folder = child.parent / device_name
target_folder.mkdir(exist_ok=True)
shutil.move(child, target_folder / child.name)
# Create a DutLib object that stores measurement data
lib_save_dir = path_to_dmt_core / "test" / "tmp" / "skywater130_lib"
lib = DutLib(save_dir=lib_save_dir, force=True)
lib.wafer = "MPW-5"
# to import all data at once, DutLib offers the import_directory method. This method needs a custom filter function to create the correct DutViews that store device-related information.
def filter_dut(dut_name):
"""Create DutView objects from the different folder names.
Parameters
----------
dut_name : str
Path to the dut folder.
Returns
-------
:class:`~DMT.core.dut_view.DutView`
DutView which should represent the device represented by a folder of measurements.
"""
dut_path = Path(dut_name)
dut_name = dut_path.parent.name
width = re.search(r"_w([\dp]+)u_", dut_path.name, re.MULTILINE).group(1)
width = float(width.replace("p", ".")) * 1e-6
length = re.search(r"_l([\dp]+)u_", dut_path.name, re.MULTILINE).group(1)
length = float(length.replace("p", ".")) * 1e-6
dut_type = DutType.n_mos if "nfet" in dut_name else DutType.p_mos
multiplication_factor = int(re.search(r"_m([\d]+)\(", dut_path.name, re.MULTILINE).group(1))
module_name = int(re.search(r"\((\d+)_", dut_path.name, re.MULTILINE).group(1))
contacts = re.search(r"_m[\d]+\((.+)", dut_path.name, re.MULTILINE).group(1)
flavor = dut_name
dut = DutMeas(
database_dir=path_to_dmt_core / "test" / "tmp",
dut_type=dut_type,
force=True,
wafer="MPW-5",
die=module_name,
width=width,
length=length,
contact_config="SGD",
name=dut_path.parent.name,
reference_node="S",
ndevices=multiplication_factor,
flavor=flavor,
)
dut.contact_info = contacts.replace("_", " ")
return dut
# To correctly assign each measurement a key in the database, the temperature_converter function is passed to import_directory for generating keys:
def key_generator(key_part):
"""The intended use is to allow different ways of temperature notice in the measurement name/key.
Each part is there for one folder level.
So usually we encounter: DEVICE_FOLDER/XXTEMPXX/measurement_name
Here this function is used to rename ALL measurements to the 2 possibilities shown in the code.
Parameters
----------
key_part : str
The part of the key which may contain the temperature
Returns
-------
float or str
Should return -1 if no temperature is obtained and otherwise the temperature in Kelvin
If a string is returned, this is used as the current key-part.
Raises
------
NotImplementedError
In case, new measurements have different structures.
"""
if "IDVG" in key_part:
return "T300.00K/IDVG"
elif "IDVD" in key_part:
return "T300.00K/IDVD"
else:
raise NotImplementedError
# Import the measurements using the dut filter function in parallel
lib.n_jobs = 6 # number of parallel jobs
lib.import_directory(
import_dir=path_to_cells,
dut_filter=filter_dut,
dut_level=2,
force=True,
temperature_converter=key_generator,
)
# Clean the data to use specifier columns
for dut_a in lib:
dut_a.clean_data()
lib.dut_ref = lib.duts[0] # does not matter currently but needed for saving and documentation
# The data is now read for further use
# to save just use:
# lib.save()
# to load in a different script:
# lib = DutLib.load(lib_save_dir)
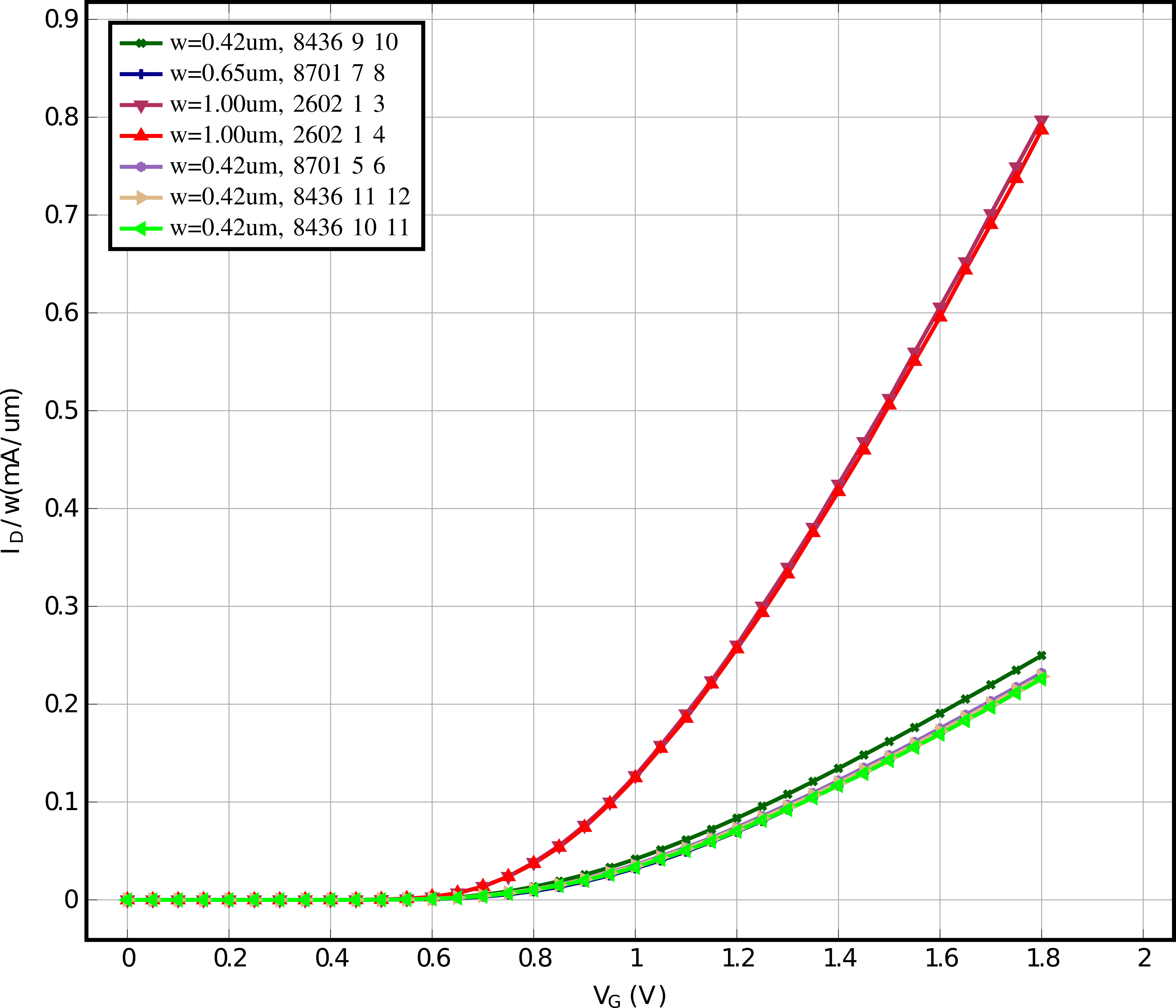
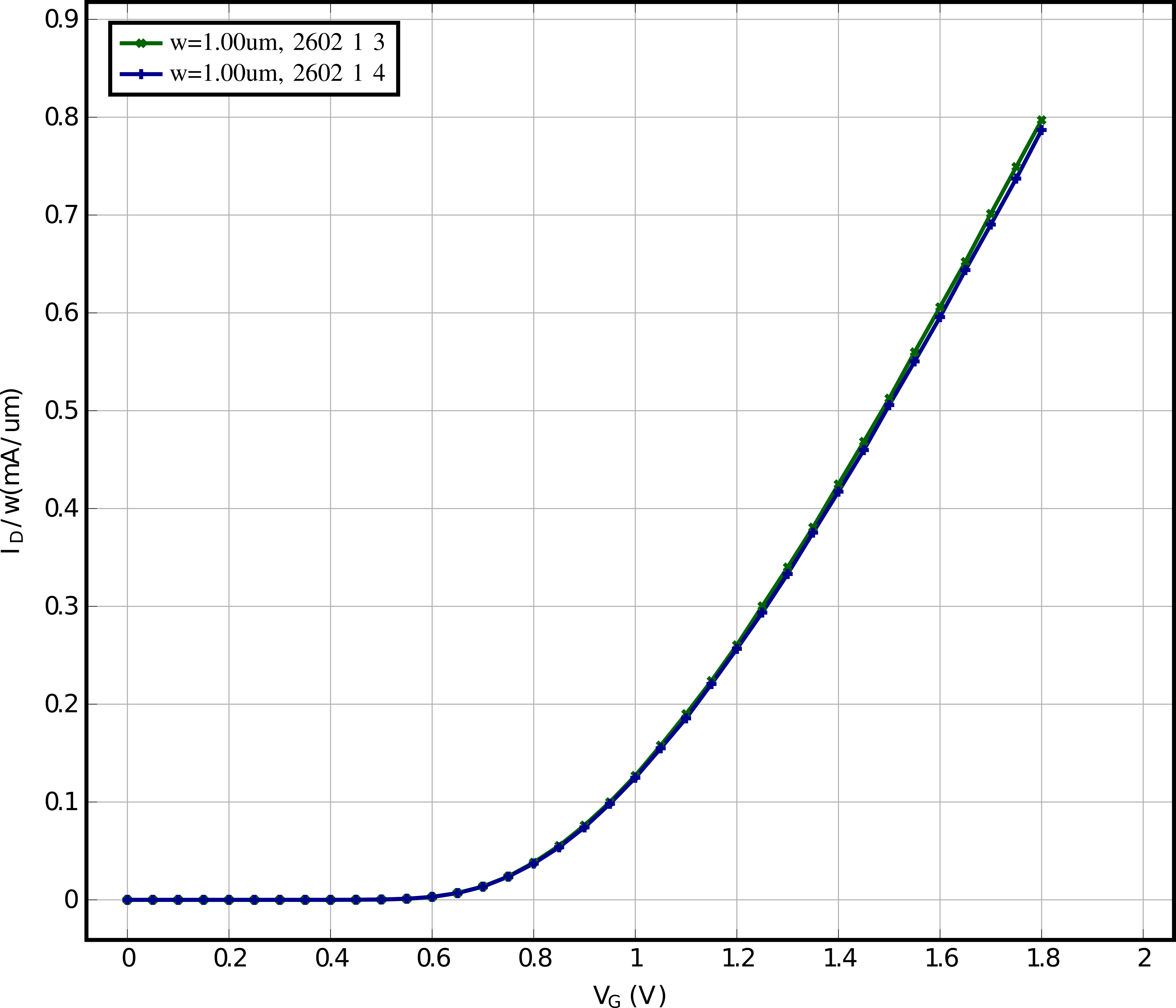
# Here we will plot JD(VG) for VB = 0, VD=1.8 for all nfet 01v8
sp_vg = specifiers.VOLTAGE + ["G"]
sp_vb = specifiers.VOLTAGE + ["B"]
sp_vd = specifiers.VOLTAGE + ["D"]
sp_ig = specifiers.CURRENT + ["G"]
sp_ib = specifiers.CURRENT + ["B"]
sp_id = specifiers.CURRENT + ["D"]
# Now we plot all data of the "nfet_01v8" devices, one plot for every unique device length
device_flavor = "nfet_01v8"
plots = []
lengths = np.unique(
[dut.length for dut in lib if dut.flavor == device_flavor]
) # to compare all lengths
lengths = [0.5e-6]
for l_i in lengths:
plot = Plot(
f"length_{l_i*1e6:.2f}um_ID_w(VG)",
x_specifier=sp_vg,
y_label="$I_{\mathrm{D}}/w(mA/um)$",
y_scale=1e3 / 1e6,
)
plot_mismatch = Plot(
f"length_{l_i*1e6:.2f}um_ID_w(VG)_mismatch",
x_specifier=sp_vg,
y_label="$I_{\mathrm{D}}/w(mA/um)$",
y_scale=1e3 / 1e6,
)
for dut in lib:
if dut.flavor == device_flavor and np.isclose(dut.length, l_i):
df = dut.data["T300.00K/IDVG"]
df = df[np.isclose(df[sp_vb], 0)]
df = df[np.isclose(df[sp_vd], 1.8)]
if dut.die in [2602, 2605, 2607, 2608, 2611, 2612, 2618, 2622, 2624, 2627]:
plot_mismatch.add_data_set(
df[sp_vg],
df[sp_id] / dut.width,
label=f"w={dut.width*1e6:.2f}um, {dut.contact_info}",
)
else:
plot.add_data_set(
df[sp_vg],
df[sp_id] / dut.width,
label=f"w={dut.width*1e6:.2f}um, {dut.contact_info}",
)
plots.append(plot)
plots.append(plot_mismatch)
for plt in plots[:-1]:
plt.plot_pyqtgraph(show=False)
plots[-1].plot_pyqtgraph(show=True)
plots[-1].save_tikz(
Path(__file__).parent.parent / "_static" / "readin_dut_lib",
standalone=True,
build=True,
clean=True,
width="6in",
legend_location="upper left",
)
# # To get a pdf with all information about the measurements DocuDutLib can be used:
# # Be aware that this feature is currently in a early state and feedback/improvemend suggestions are very welcome.
# docu = DocuDutLib(lib, devices=[{"name": "esd_nfet_01v8"}])
# docu.generate_docu(
# lib_save_dir.parent / "docu_skywater130_raw",
# plot_specs=[
# {"type": "id(vg)", "key": "IDVG", "dut_type": DutType.n_mos},
# {"type": "id(vg)", "key": "IDVG", "dut_type": DutType.p_mos},
# {"type": "id(vd)", "key": "IDVD", "dut_type": DutType.n_mos},
# {"type": "id(vd)", "key": "IDVD", "dut_type": DutType.p_mos},
# ],
# show=False,
# save_tikz_settings={
# "width": "3in",
# "height": "5in",
# "standalone": True,
# "svg": False,
# "build": True,
# "mark_repeat": 20,
# "clean": True, # Remove all files except *.pdf files in plots
# },
# )
The generated files